Latest Projects
Network Discovery
We use computational approaches to generate hypotheses about microbial networks and design experiments to test our understanding of these networks. Read More >>>
Metabolic Engineering
We develop computational approaches to predict how different genetic perturbations impact cellular production and growth rates. Read More >>>
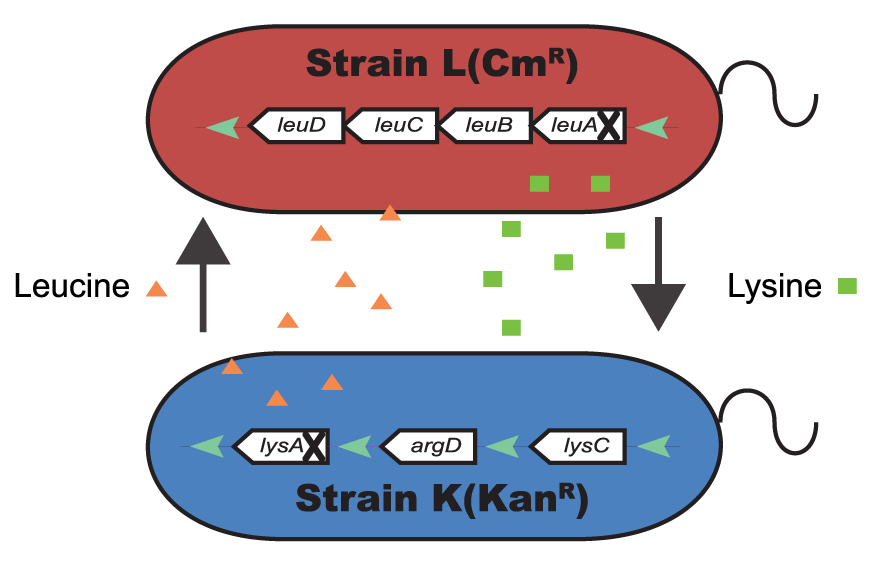
Microbial Interactions
We use systems biology approaches to study microbe-microbe and/or microbe-host interactions and develop models and approaches to predict metabolic fluxes and metabolic interactions in these complex systems. Read More >>>
Welcome to Reed Laboratories!
Our group studies microbial metabolism and regulation using a combination of computational and experimental approaches. Overall, we use computational models to study biological systems, engineer cells, and expand our knowledge of the underlying mechanisms behind observed cellular behaviors.
